We seek to understand the underlying principles of molecular recognition and specificity for macromolecular complexes involved in misfolding diseases. To achieve this we develop strategies that integrate mass spectrometry with structural and computational approaches to understand the structure function relationship of protein machinery involved in protein misfolding pathways. How protein-based molecular recognition drives protein misfolding diseases is an important fundamental question in understanding the mechanisms underlying neurodegenerative diseases.
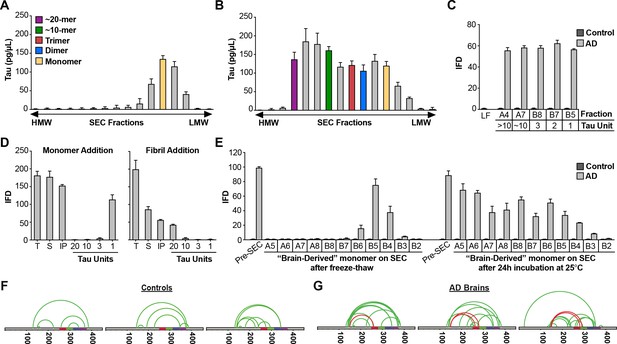
AD brain contains seed-competent monomer.
Tau from control and AD brains was immunoprecipitated and subjected to SEC. (A) SEC from control brain contained predominantly tau monomer. (B) SEC from AD brain contained a range of tau assembly sizes. (C) Tau monomer from control brain exhibited no seeding activity, whereas monomer from AD brain did, along with larger assemblies (p<0.001). Tau Unit refers to the putative number of molecules per assembly. LF = Lipofectamine control. (D) Tau KO mouse brain was spiked either with human tau Ms or fibrils prior to dounce homogenization, immunopurification, and resolution by SEC. Samples spiked with Ms exhibited monomer seeding activity, but not samples that had been spiked with fibrils. (E) AD-derived tau monomer was incubated for the indicated times prior to SEC and determination of seeding activity in each fraction. Larger seed-competent assemblies formed after 24 hr incubation at RT. (F, G) Three control and AD brains were homogenized, monomer isolated, and evaluated by XL-MS. Tau monomer from controls lacked the long-range crosslinks observed in Ms. AD-derived Ms contained long-range crosslinks (aa150 to aa254-290) also observed in recombinant forms of Ms.
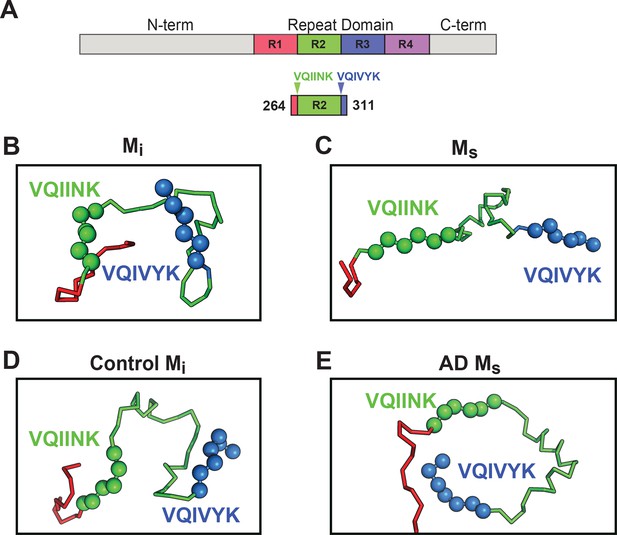
Models of Mi and Ms suggest differences in the R1R2 and R2R3 regions.
XL-MS identified pairs were used as restraints in Rosetta to create structural models of discrete tau domains. (A) Schematic highlighting the region of the RD encoding structural differences between Mi and Ms. Tau RD is colored in red (R1), green (R2), blue (R3) and indigo (R4); N- and C-terminal portions of tau are shown in grey. Fragments of interest are shown with their position in the RD. (B) recombinant Mi; (C) fibril-derived Ms, (D) Control Mi and (E) AD-derived Ms. Regions surrounding the R1R2 and R2R3 are indicated, highlighting two amyloid-forming sequences, VQIINK (green spheres) and VQIVYK (blue spheres). In both forms of Mi VQIINK and VQIVYK are associated with flanking amino acids in hairpin structures. In both forms of Ms the VQIINK and VQIVYK sequences are presented at the protein surface.